Energy funcion for searching global minimum
- Parameters:
-
ndim dimension of the parametric space (usually 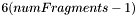
X vector from parametric space E value of the energetic funciton result value if error flag
- See also:
- InitComputation()
#include <ReadData.h>
Public Member Functions | |
| CData () | |
| ~CData () | |
| void | Read (char *name) |
| void | Delete () |
| void | PrintSelf () |
| void | Prepare () |
| void | Postpare () |
| void | CheckAllRestraints () |
| int | GetNumber (int id, int *idFrg, int *idLoc) |
| void | EnergyFunction (int ndim, const ColumnVector &X, REAL &E, int &result) |
| void | InitComputation (int ndim, ColumnVector &x) |
| void | SetFilenamePDBall (CSmallString &str) |
| void | SetFilenamePDBfinal (CSmallString &str) |
| CSmallString & | GetFilenamePDBall () |
| CSmallString & | GetFilenamePDBfinal () |
| void | SetFilenameEnergyGraph (CSmallString &str) |
| CSmallString & | GetFilenameEnergyGraph () |
| void | ApplyTransformsToAllAtoms (const ColumnVector &X) |
| void | ExportDataPDB (const ColumnVector &X, FILE *fout) |
| void | ExportDataPDB (const ColumnVector &X, char *name) |
| void | ExportDataXML (const ColumnVector &X, char *name) |
| void | SetNumIterations (int a) |
| void | IncrementNumIterations (int a=1) |
Public Attributes | |
| int | numFragments |
| int * | nF |
| CSmallString * | idF |
| int | masterFragment |
| Atom ** | Atoms |
| int ** | changingAtoms |
| int * | cntChangeAtom |
| Point ** | points |
| Point * | centerF |
| REAL * | chargeF |
| bool * | distribF |
| int | numRestrains |
| CRestrain ** | restrains |
| CContrELECT * | contrEl |
| CSmallString | strFilePDBall |
| CSmallString | strFilePDBfinal |
| CSmallString | strFileEnergyGraph |
| int | numIterations |
|
|
construcor |
|
|
destrucor |
|
|
Applies transformations driven by vector X to all atoms in fragments
|
|
|
Function check if the restraints are well defined if yes the computation goes on otherwise the program ends |
|
|
Release memory |
|
||||||||||||||||||||
|
Energy funcion for searching global minimum
|
|
||||||||||||
|
Export data to PDB file
|
|
||||||||||||
|
Export data to PDB file
|
|
||||||||||||
|
Export data to XML file
|
|
|
Returns filename to save energy graph |
|
|
Returns filename to save all frames of computation to PDB-file |
|
|
Returns filename to save final (optimized) constitution |
|
||||||||||||||||
|
Get number of fragment and position of atom in this fragment
|
|
||||||||||||
|
Initial setting for optimalization procedure
|
|
|
Function called after optimalization |
|
|
Prepare data for optimalization (creates some temporary tables) |
|
|
Print Information and content of this class |
|
|
Reads data from XML file
<?xml version="1.0" encoding="ISO-8859-1"?> <!-- testovaci soubor s daty //--> <FIT> <FRAGMENTS> <FRAGMENT id="F1" elementarcharge="1" distribute="no"> <ATOM x="-2" y="0" z="0" type="H" id="1" /> <ATOM x="0" y="0" z="0" type="O" id="2"/> <ATOM x="2" y="0" z="0" type="H" id="3"/> </FRAGMENT> <FRAGMENT id="F2" master="yes" elementarcharge="1" distribute="no"> <ATOM x="10" y="-2" z="-6" type="H" id="4"/> <ATOM x="10" y="0" z="-6" type="C" id="5"/> <ATOM x="10" y="2" z="-6" type="H" id="6"/> </FRAGMENT> </FRAGMENTS> <RESTRAINTS> <RESTRAINT type="DIST" part1="2" part2="5" value="5" k="1" /> <RESTRAINT type="ANGLE" part1="1" part2="2" part3="2" part4="5" value="1.57" k="1" disabled="yes" /> <RESTRAINT type="TORSION" part1="1" part2="2" part3="5" part4="6" value="1.57" k="100" disabled="no"/> <RESTRAINT type="ELECTROSTATIC" disabled="no" k="10" eps="0.1"/> </RESTRAINTS> </FIT> |
|
|
Set filename for energy graph |
|
|
Set filename for PDB output for sequence |
|
|
Set filename for PDB output for final constitution |
 1.3.4
1.3.4